A. Gangi1, W. Sun1, S. Yoder3, M. Fournier2, M. C. Lee1 1Moffitt Cancer Center,Comprehensive Breast Program,Tampa, FL, USA 2Moffitt Cancer Center,Tissue Core Facility,Tampa, FL, USA 3Moffitt Cancer Center,Molecular Genomics Core Facility,Tampa, FL, USA
Introduction: Translational cancer research is increasingly reliant on existing tissue and data banks for retrospective tissue and data studies with potential clinical impact. Despite significant infrastructure and funding, these investigations are hindered by a multitude of unanticipated hurdles.
Methods: This is a funded, prospective/retrospective, IRB-approved, tissue and data study incorporating SNP analysis of breast cancer patients treated at an NCI designated, comprehensive cancer center. All cases and controls had archival tissue and were at least 5 years from their incident breast cancer diagnosis. Large-scale prospective institutional and programmatic databanks, tissue banks, and medical record chart review were leveraged. Tissue specimens were reviewed by a research pathologist for verification and adequacy. Sequencing of 10 selected genes at 30x coverage was targeted. Causes of attrition in the 1:1 matched case-control population were evaluated. Matched pairs were not matched by tissue type.
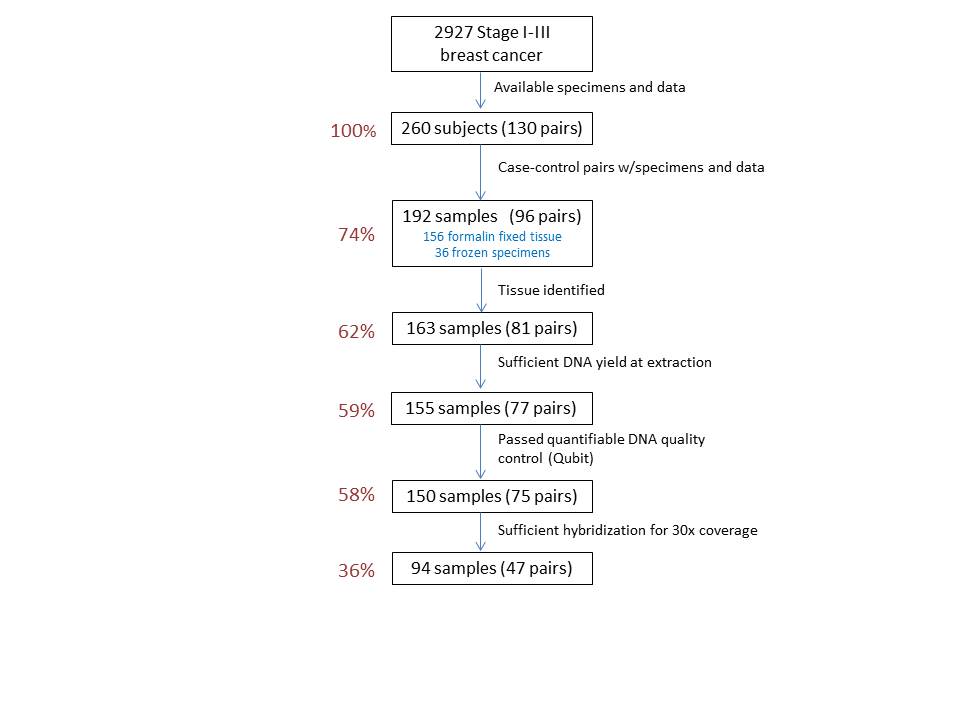
Results: 2927 stage I-III breast cancer patients were reviewed to identify 130 matched case-control pairs (260 total subjects) with specimens and data. Because of paired matching, elimination of one specimen resulted in the elimination of a pair if a replacement could not be identified. Of 260 specimens (130 pairs), 192 tissue samples (96 matched pairs) were identified (73.8%) over an 18-month time frame. 156 were archived FFPE (slides or blocks) and 36 were frozen (solid tissue, blood, extracted DNA). Thirty-four of the 130 data-matched pairs were unusable (26.1%): in 28 pairs, tissue could not be identified for one of the paired subjects. This was due to: missing blocks/slides, compromised tissue cases, or specimens that did not represent normal tissue due to inflammation or necrosis. In 2 other pairs, adequate tissue for macrodissection could not be identified. Four additional pairs were excluded due to insufficient DNA yields at extraction for one of the paired specimens. Of 192 specimens, 150 (75 pairs) passed the quantifiable DNA quality control studies (Qubit). Of 75 pairs sent for DNA sequencing, 47 pairs (36.2%) had evaluable data. The remaining 28 had insufficient hybridization for 30x coverage.
Conclusion: Despite significant infrastructure and resources, retrospective tissue studies are fraught with evaluable specimen loss at each step of the process. When assessing feasibility of retrospective tissue studies, investigators should consider a significant dropout rate within populations of archival tissue specimens.