A. L. Moore1,3, C. Marshall1, U. Litzenburger4, R. C. Ransom4, L. Barnes4, B. Duoto4, D. Foster1, R. E. Jones2,4, S. Mascharak4, M. Hu4, H. Y. Chang4, H. P. Lorenz1, M. T. Longaker1 1Stanford University,Department Of Surgery,Palo Alto, CA, USA 2University Of Texas Southwestern Medical Center,Department Of Surgery,Dallas, TX, USA 3Brigham And Women’s Hospital,Department Of Surgery,Boston, MA, USA 4Stanford University,Palo Alto, CA, USA
Introduction:
Skin scarring poses a major financial and physiologic burden on patients and the US healthcare system. In 2015, our group discovered a subset of fibroblasts in the dorsal dermis identified by embryonic expression of the homeobox transcription factor Engrailed-1 that deposit the bulk of scar collagen in adult mice. These cells are also present in embryonic development around the time of phenotypic transition from regenerative scarless healing to fibrotic scarring. Given Engrailed-1 positive fibroblasts (EPFs) share a common precursor cell, we hypothesized that the EPFs accumulate epigenetic changes over time that result in their phenotypic change and result in a permanent cellular phenotype.
Methods:
Fibroblasts were isolated from En1Cre; R26mTmG mice using fluorescence-activated cell sorting (FACS) at gestational ages embryonic day (e)10, e16, e18, postnatal day (p)1, p30, and p30 wounded skin. Epigenetic regulation was analyzed using the assay for transposase accessible chromatin using sequencing (ATAC-seq). Fibroblasts were then transplanted from e16 embryos to p1 mouse pups, and vice versa, to observe scar forming behavior in vivo.
Results:
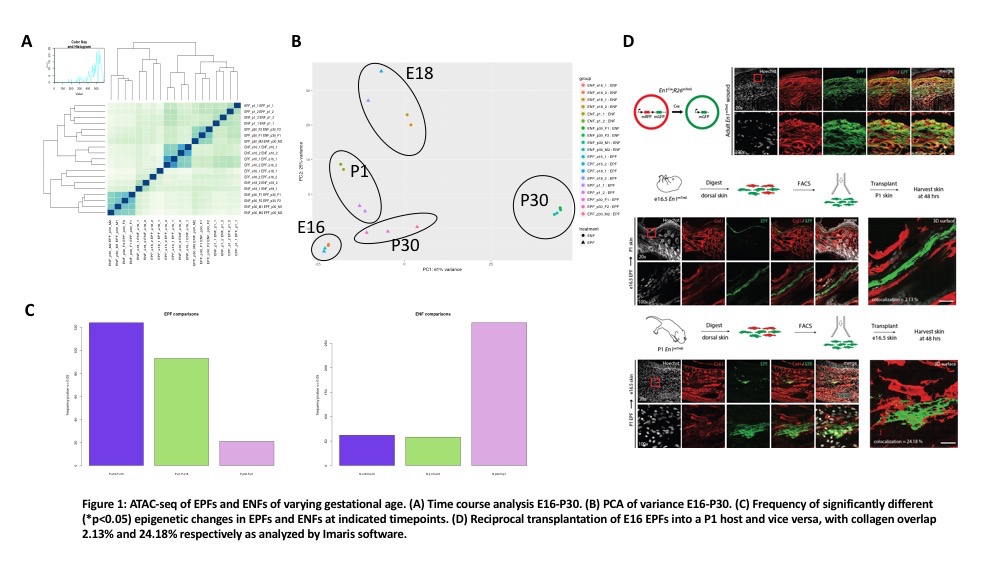
E10 fibroblasts are of a single lineage and were excluded from analysis. Time course analysis of e16-p30 EPFs and Engrailed-1 negative fibroblasts (ENFs) shows appropriate correlation between samples (Figure 1A). Grouping analysis of variance shows p30 EPFs and ENFs being the most dissimilar, and EPFs from p30 are most like e16 EPFs (Figure 1B). Most epigenetic changes in the EPF lineage occur in embryonic development between e16 and e18 with fewer epigenetic changes occurring postnatally between p1 and p30 (significant peaks = 124 vs. 20, Figure 1C). In contrast, the ENF lineage accumulates more epigenetic changes between p1 and p30 (significant peaks = 62 vs. >250, Figure 1C). Lastly, reciprocal transplantation of e16 fibroblasts into a p1 host and vice versa reveal a significant difference in collagen overlap (2.13% versus 24.18%) and morphologic changes suggestive of quiescence versus reactivity (Figure 1D).
Conclusions:
ATAC-seq analysis of EPF lineage cells reveals most epigenetic changes occurring at the time of phenotypic change from scarless to scarring (between e16 to e18). In future experiments, we will identify transcription factors to perturb using CRISPR-Cas9. Manipulated cells will be transplanted into fetal and postnatal hosts. These experiments will reveal if master regulators are involved in the scar forming phenotype of EPFs.